11. BindingSite Analysis:
Protein Function
Protein function
-
A protein is not a standalone system.
-
The coordinated action of several proteins controls how cells function.
-
Ligands, such as tiny chemical compounds, interact with proteins, nucleic acids, and other proteins.
3D-Figure 1.
PDB structure of 5EMC
is of Transcription factor GRDBD and smGRE complex with Two chains
B [auth A],
C [auth B] of sequence length 94
(CLICK me for
more info!!).
3D-Figure 2. PDB structure of 4UDC is of GR in complex with dexamethasone with chain A of sequence length 280 (CLICK me for more info!!).
Binding site
Defined portion of the protein surface involved in the interaction with a ligand or a macromolecule.
Intermolecular Interactions
-
Van der Waals forces
-
Hydrogen bonds
-
Salt bridges
-
Hydrophobic effect
Diversity of binding sites
Proteins interact with:
-
Proteins
-
Nucleic acids
-
Ligands
Different type of interactions require different binding sites
Below 3D interactive view gives us a model of :
-
PDB ID: 4DXA
Rapl in complex with KRIT1
Res: 1.95 A
Ligand binding site: deep cleft
PPI: flat and large surface
Recognition determinants
-
Geometrical complementarity (Size and shape)
-
Physicochemical complementarity (specific interactions)
-
Pocket volume: 600-900 Å3
Drug-like molecule volume: 400 Å3 -
Specific amino acids occur more frequently in BindSites (R, H, W, Y)
Protein flexibility
Flexibility of the binding site may result from:
-
rotating the side chains
-
structural reorganization (backbone movement)
To accommodate various ligands, the binding site may need to adopt a varied conformation (induced fit).
Where performing binding site analysis, it is desirable to take into account various 3D protein structures (when available).
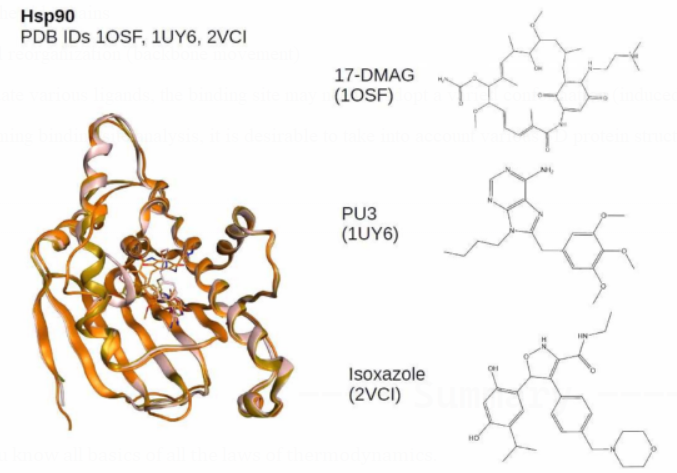
Binding site identification
A co-crystallized ligand can quickly locate a protein's binding site.
-
What if there are no co-crystallized ligands available?
-
What happens if we want to investigate brand-new binding sites (allosteric sites)?
It has become possible to locate binding sites inside a protein structure using computational methods.
two divisions:
-
Algorithms based on geometry
-
Energy-efficient algorithms
Algorithms based on
geometry
Use just geometric criteria to identify solvent-accessible areas
embedded in the protein surface. virtual sphere fitting (e.g. Site
Finder)
Algorithms based on energy
Calculate the energy of the interaction between a probe and a protein.
Druggability
What qualifies a protein as a viable therapeutic target?
-
The protein and the relevant illness are related.
-
The binding of a tiny chemical can alter how biologically active a protein is.
How to tell if a protein can be drugged:
-
Find a binding pocket, then check to see if it can bind compounds similar to drugs.
What makes a
BindingSite druggable?
physical, chemical, and structural characteristics that encourage the
binding of tiny molecules with high specificity and affinities.
the following factors: size, shape, and flexibility.
Define a BindingSite's druggability by contributing.
What elements
influence druggability?
The interaction of various parameters leads to druggability.
Indicators that positively affect druggability include:
- Buriedness - Hydrophobicity - Pocket size
It has been proposed that the polar contact area has a deleterious
impact.
For drug binding, polar interactions are still very important! (Salt
bridges and H-bonds)
---- Summary ----
As of now you know all basics of all the laws of thermodynamics.
-
Protein function
-
CFactors influencing druggability
-
Druggability
-
etc..
________________________________________________________________________________________________________________________________

________________________________________________________________________________________________________________________________
